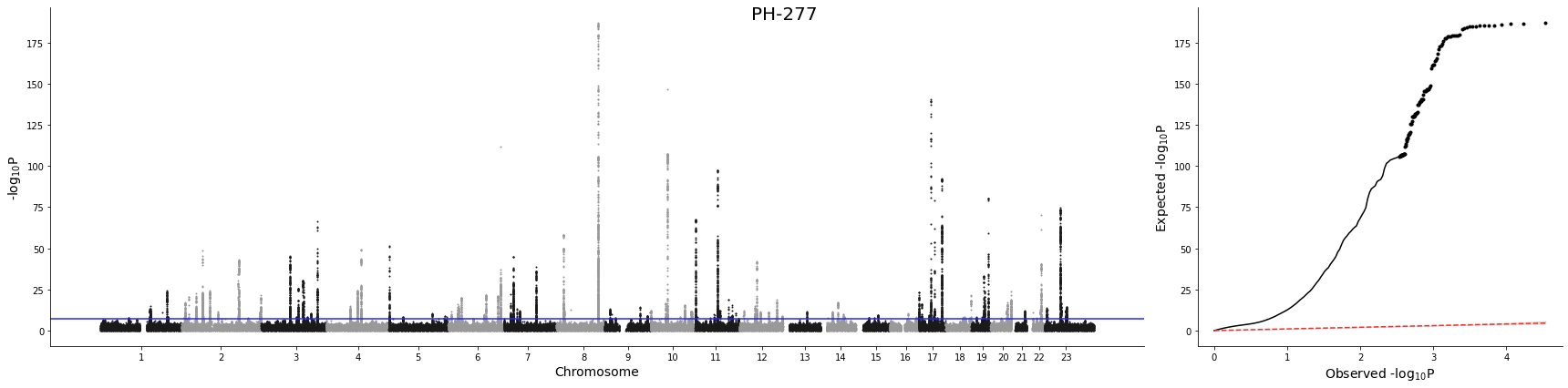
Manhattan & QQ plot¶
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib import gridspec
from scipy.stats.mstats import mquantiles
from scipy.stats import beta
from scipy.stats import linregress
sum_stat = '/f/jianhua/nankai-hic/fine-mapping/prostate_cancer.txt'
chrom, bp, p = 'CHR','BP','P'
sep = '\t'
df = pd.read_csv(sum_stat,sep=sep)
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
<ipython-input-2-949a920fa523> in <module>
2 chrom, bp, p = 'CHR','BP','P'
3 sep = '\t'
----> 4 df = pd.read_csv(sum_stat,sep=sep)
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in read_csv(filepath_or_buffer, sep, delimiter, header, names, index_col, usecols, squeeze, prefix, mangle_dupe_cols, dtype, engine, converters, true_values, false_values, skipinitialspace, skiprows, skipfooter, nrows, na_values, keep_default_na, na_filter, verbose, skip_blank_lines, parse_dates, infer_datetime_format, keep_date_col, date_parser, dayfirst, cache_dates, iterator, chunksize, compression, thousands, decimal, lineterminator, quotechar, quoting, doublequote, escapechar, comment, encoding, dialect, error_bad_lines, warn_bad_lines, delim_whitespace, low_memory, memory_map, float_precision, storage_options)
608 kwds.update(kwds_defaults)
609
--> 610 return _read(filepath_or_buffer, kwds)
611
612
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in _read(filepath_or_buffer, kwds)
460
461 # Create the parser.
--> 462 parser = TextFileReader(filepath_or_buffer, **kwds)
463
464 if chunksize or iterator:
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in __init__(self, f, engine, **kwds)
817 self.options["has_index_names"] = kwds["has_index_names"]
818
--> 819 self._engine = self._make_engine(self.engine)
820
821 def close(self):
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in _make_engine(self, engine)
1048 )
1049 # error: Too many arguments for "ParserBase"
-> 1050 return mapping[engine](self.f, **self.options) # type: ignore[call-arg]
1051
1052 def _failover_to_python(self):
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in __init__(self, src, **kwds)
1865
1866 # open handles
-> 1867 self._open_handles(src, kwds)
1868 assert self.handles is not None
1869 for key in ("storage_options", "encoding", "memory_map", "compression"):
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/parsers.py in _open_handles(self, src, kwds)
1360 Let the readers open IOHanldes after they are done with their potential raises.
1361 """
-> 1362 self.handles = get_handle(
1363 src,
1364 "r",
~/anaconda3/envs/jb-book/lib/python3.8/site-packages/pandas/io/common.py in get_handle(path_or_buf, mode, encoding, compression, memory_map, is_text, errors, storage_options)
640 errors = "replace"
641 # Encoding
--> 642 handle = open(
643 handle,
644 ioargs.mode,
FileNotFoundError: [Errno 2] No such file or directory: '/f/jianhua/nankai-hic/fine-mapping/prostate_cancer.txt'
Reduce size¶
df['CHR'] = df['CHR'].replace('X',23)
df['CHR'] = df['CHR'].astype(int)
df['P'] = -np.log10(df['P'])
df['P_down'] = df['P'].round(1)
df['P'] = df['P'].round(3)
df['BP_down'] = df['BP']/5e6
df['BP_down'] = df['BP_down'].astype(int)
idx = np.random.permutation(np.arange(len(df)))
df = df.iloc[idx].drop_duplicates(['CHR','P_down','BP_down'])
df = df.sort_values(['CHR','BP'])
df['P'] = 10 ** -df['P']
def qq(data,ax,color):
xmax = 0
ymax = 0
alpha = 0.9
color = '#000000'
n_quantiles = 100
q_pos = np.concatenate([
np.arange(99.) / len(data),
np.logspace(-np.log10(len(data)) + 2, 0, n_quantiles)
])
q_data = mquantiles(data, prob=q_pos, alphap=0, betap=1, limit=(0, 1))
q_th = q_pos.copy()
q_err = np.zeros([len(q_pos), 2])
for i in range(0, len(q_pos)):
q_err[i, :] = q_err[i, :] = beta.interval(
alpha,
len(data) * q_pos[i],
len(data) - len(data) * q_pos[i])
q_err[i, q_err[i, :] < 0] = 1e-15
slope, intercept, r_value, p_value, std_err = linregress(q_th, q_data)
xmax = np.max([xmax, -np.log10(q_th[1])])
ymax = np.max([ymax, -np.log10(q_data[0])])
ax.plot(
-np.log10(q_th[n_quantiles - 1:]),
-np.log10(q_data[n_quantiles - 1:]),
'-',
color=color)
ax.plot(
-np.log10(q_th[:n_quantiles]),
-np.log10(q_data[:n_quantiles]),
'.',
color=color,
label='gf')
ax.plot([0, xmax], [0, xmax], '--k',color='#f42e30')
ax.fill_between(
-np.log10(q_th),
-np.log10(q_err[:, 0]),
-np.log10(q_err[:, 1]),
color=color,
alpha=0.1,
)
def manhattan(df,ax):
df[p] = -np.log10(df[p])
df = df.sort_values(chrom)
df_grouped = df.groupby((chrom))
colors = ['#1A1A1A','#999999',]
x_labels = []
x_labels_pos = []
end = 1000
for num, (name, group) in enumerate(df_grouped):
group[bp] = group[bp] + end
end = group[bp].max() + 1000
ax.scatter(group[bp], group[p],c=colors[num % len(colors)],s=1)
x_labels.append(name)
x_labels_pos.append(group[bp].mean())
ax.axhline(y=-np.log10(5e-8), color='#2222FF', linestyle='-')
ax.set_xticks(x_labels_pos)
ax.set_xticklabels(x_labels)
# print(df.loc[0,bp]-len(df)*0.1,end+len(df)*0.1)
# ax.set_ylim([-0.5, df[p].max()*1.05])
# df = alldf.copy()
figure_tile = 'PH-277'
fig = plt.figure(figsize=(24, 6))
gs = gridspec.GridSpec(1, 2, width_ratios=[3, 1])
ax0 = plt.subplot(gs[0])
ax1 = plt.subplot(gs[1])
qq(df[p], ax1, 'b')
manhattan(df,ax0)
# ax0.set_xlim(left=-3e7,right=2.9e9)
ax0.set_xlabel('Chromosome', fontsize=14)
ax0.set_ylabel('-$\mathregular{log_{10}}$P', fontsize=14)
ax1.set_xlabel('Observed -$\mathregular{log_{10}}$P', fontsize=14)
ax1.set_ylabel('Expected -$\mathregular{log_{10}}$P', fontsize=14)
ax0.spines['right'].set_visible(False)
ax0.spines['top'].set_visible(False)
ax1.spines['right'].set_visible(False)
ax1.spines['top'].set_visible(False)
fig.suptitle(figure_tile, fontsize=20)
fig.tight_layout()
fig.savefig('{}_Manhattan_QQ.pdf'.format(figure_tile), dpi=300)
/f/jianhua/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:22: RuntimeWarning: invalid value encountered in less
/f/jianhua/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:33: RuntimeWarning: divide by zero encountered in log10
/f/jianhua/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:40: RuntimeWarning: divide by zero encountered in log10
/f/jianhua/anaconda3/lib/python3.7/site-packages/ipykernel_launcher.py:11: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
# This is added back by InteractiveShellApp.init_path()